[ad_1]
In the past I’ve written about text matching with dictionaries using David Smiley’s SolrTextTagger project. SolrTextTagger uses Lucene’s finite state transformer (FST) technology to implement a finite state machine (FSM) used by the Aho-Corasick string matching algorithm. As stated on its Wikipedia page, the Aho-Corasick algorithm has good performance properties that make it very attractive for text annotation, which is also an interesting use case. To support the use case, I created a Solr Dictionary Annotator (SoDA) project that wraps the SolrTextTagger service in a slightly more usable API and provides multiple levels of matching (exact, minor, stem, etc.).
To set the SolrTextTagger index, or the Aho-Corasick string matching algorithm in general, the basic idea is to populate the FSM with dictionary entries. Internally, this will create a Trie structure that associates the words in the dictionary entries with each other. At runtime, you would pass text against this structure and it would match the text phrases that match the dictionary entries. One major disadvantage of this approach is that it only matches phrases in the text that are in the same order as the dictionary equivalent phrase. For example, if my dictionary entry is “pulmonary tuberculosis”, it does not match “pulmonary tuberculosis” in the text. Depending on your application, this may or may not be important. But I recently discovered a way to do this, thanks to ideas presented in the paper Effective Clinical Concept Extraction in Electronic Medical Records (Guo, Kakrania, Baldwin, and Syeda-Mahmood, IBM Research Almaden, 2017). Hat tip to my manager Ron Daniels for getting me on paper.
Specifically, the part that caught my eye was their example of being able to match the string “CT of the chest” from the phrase “post CT scan of the chest” and the description of their approach in the paper. As you can see, there’s not much detail there, so I’m not sure if my approach is identical to theirs, but it got me thinking and resulted in the algorithm I describe in this post. In the spirit of reproducibility, the description is accompanied by my Jupyter notebook, which contains the code for building the FSM and using the FSM based on it using the PyAhoCorasick library. The notebook contains several examples of using the Aho-Corasick algorithm to match strings against an internal medical dictionary, described in an imprecise manner, for short queries (including the example in the paper) as well as long paragraph texts.
To speed up the processing, we propose a new indexing method that significantly reduces the search space and at the same time maintains the corresponding flexibility. First, each word in the dictionary is represented by a unique prefix, the shortest sequence of letters that distinguishes it from all other terms. Next, an inverted index is created from the prefixes to the propositional mappings. Starting with a representative prefix for each term in the dictionary (or a set of prefixes in the case of a multi-word term), all relevant sentences can be easily searched as potential matches for the term and further filtered by the longest common word. Sequence matching can be used to further refine search results.
The big idea here is the following. Instead of populating the FSM dictionary with phrases, we populate it with words from which phrases are built. When we filled it with phrases, the payload associated with it was the concept ID. When we fill it with words, the chance of collision is almost certain. For example, the word “tuberculosis” is found 1352 times in the dictionary. So the payload is now a list of concept ID and synonym ID pairs, as well as the inverted index structure used by search engines. A concept ID is self-defining, a synonym ID is a running serial number (for example, synonym ID 0 is the base name of the concept). We filter out stop words and non-alpha words to reduce noise. Matches are not case sensitive (both dictionary and text are lower case), unless the word is an obvious abbreviation and is specified in all caps.
In addition to the concept synonym ID pair (CSID hereafter), we also store in the payload a floating point number representing the match value. The idea is that the string “tuberculosis” corresponding to the concept “tuberculosis” is more valuable than the string “pulmonary tuberculosis” or “pulmonary tuberculosis symptoms” against the concept. The weight of each term in a synonym is the reciprocal of the number of words in the synonym (minus terms). This will later be used to evaluate the quality of the matches. Thus, the payload of each term is the list of pairs (CSID, weight) for each occurrence of the term in the synonym.
Since the PyAhoCorasick API does not allow us to incrementally add the payload, we preprocess a TSV file representing a dictionary to create a dictionary of terms and a list of (CSID, weight) pairs. We then use the dictionary to populate the Aho-Corasick FSM. Internally, FSM states are just symbols, so when you query this structure, you have to filter the output, keeping matches that match the query terms.
When querying, you should use the same transformations as you did for dictionary terms, namely, except for lower case, unless there is an obvious abbreviation, remove the terms and non-alpha words. As mentioned earlier, you need to filter the results to keep only matches with full terms. The next step is to further process the results to remove the set of inexact matches.
The first step is to populate the weight matrix W using the payload weights of the filtered results. The rows of this matrix correspond to the number of matching terms, and the columns correspond to the number of CSID entries in all terms. As the terms cluster around the CSID, the matrix is expected to be relatively sparse. The degree to which the entire query string matches a particular CSID is evaluated as the sum of the corresponding column weights. You provide a confidence limit that the algorithm will use to only retain matches whose row sum is greater than it. These are now moved to the candidate matching matrix C. The figure below shows the offsets and weights returned for the various CSIDs for the query “helicobacter pylori patient perception”.
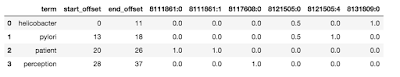
Here are some interesting things to note. Notice that both “Helicobacter” and “Pylori” have their own matches (8131809:0 and 8121505:4). Additionally, both matched CSID 8121505:0. The term “patient” exactly matches two CSIDs — 811861:0 and 811861:1. This is because the term is repeated in synonyms, one with a capital P and one without. However, there may be cases where the same term may refer to many different concept IDs because they are truly ambiguous.
The next step is to combine the offsets. This is where we take terms that are consistent or close to each other and treat them as a single span. The proximity is determined by the user-specified span_slop, which is set to 5 by default. In our example above, the first two terms can be combined for CSID 8121505:0. At this point, we have moved from annotating a single term to multiple CSID pairs as well as annotating multiple terms.
We now open maps that are entirely contained within longer maps. The idea here is that a string like “Helicobacter pylori” is more specific than “Helicobacter pylori” or “pylori”, and the latter maps to the former in favor of the former. So this is what the algorithm does next. Now we are ready to enter the relevant concept base name and present the final results as shown below.
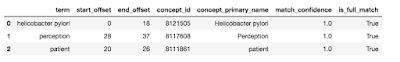
And that’s quite a lot. This technique applies not only to short strings like I used in the example, but also to longer pieces of text. In my notebook with code and examples, I show the same algorithm used to map entire paragraphs, although for quality reasons it may be better to annotate sentence by sentence.
At some point, I hope to put this functionality into SoDA, my open source wrapper for SolrTextTagger that I mentioned earlier. But given my current queue, I don’t foresee that happening anytime soon. If any of you would like to implement this idea in Scala and integrate it into SoDA, please send me a request and I’ll be happy to integrate and give you credit.
[ad_2]
Source link