[ad_1]
AllenAI’s SciSpacy project provides a language model trained on biomedical text that can be used for named entity recognition (NER) of biomedical entities using the standard SpaCy API. Unlike entities found using SpaCy’s language models (at least English), where entities have types such as PER, GEO, ORG, etc., SciSpacy entities have a single type of ENTITY. For their further classification, SciSpacy provides Entity Linking (NEL) functionality through its integration with various ontology providers such as Unified Medical Language System (UMLS), Medical Subject Headings (MeSH), RxNorm, Gene Ontology (GO) and Human Phenotype Ontology (HPO) .
NER and NEL processes are separated. The NER process finds candidate entity boundaries and matches these ranges to corresponding ontologies, which may result in zero or more ontology entries corresponding to the range. All candidate ranges are then matched against all relevant entities.
In this post, I’ll describe a strategy related to the uncertainty of entities. Based on limited testing, this selects the correct concept about 73% of the time.
The strategy is based on the intuition that an ambiguously related unit range is more likely to resolve a concept that is closely related to the concepts of other unambiguous units in the sentence. In other words, the best target label for an ambiguous entity is the one that is semantically closest to the labels of the other entities in the sentence. Or even more succinctly, and with apologies to John Firth, a unit is known by the company it keeps.
The fact that viral antigens cannot be demonstrated is used coloring There is no result Antibodies is present cat that is already attached to these antigens and prevents It is mandatory of others Antibodies.
The NEL step will attempt to link these spans to the UMLS ontology. The matching results are shown below. As mentioned earlier, each UMLS concept maps to one or more schematic types, and these are also shown here.
| Subject-ID | unit span | Concept-ID | The basic name of the concept | Semantic type code | Semantic type name |
|---|---|---|---|---|---|
| 1 | coloring | C0487602 | coloring method | T059 | laboratory procedure |
| 2 | Antibodies | C0003241 | Antibodies | T116 | amino acid, peptide or protein |
| T129 | immunological factor | ||||
| 3 | cat | C0007450 | Felis catus | T015 | A mammal |
| C0008169 | Chloramphenicol O-acetyltransferase | T116 | amino acid, peptide or protein | ||
| T126 | Enzyme | ||||
| C0325089 | Felidae family | T015 | A mammal | ||
| C1366498 | chloramphenicol acetyl transferase gene | T028 | A gene or genome | ||
| 4 | antigens | C0003320 | antigens | T129 | immunological factor |
| 5 | Mandatory | C1145667 | Mandatory action | T052 | activity |
| C1167622 | binding (molecular function) | T044 | molecular function | ||
| 6 | Antibodies | C0003241 | Antibodies | T116 | amino acid, peptide or protein |
| T129 | immunological factor |
A sequence of entity spans, each mapped to one or more semantic type codes, can be represented by a graph of semantic type nodes, as shown below. Here, each vertical grouping corresponds to a unit position. A BOS node is a special node that represents the beginning of a sequence. From our intuition above, unit uncertainty is now just finding the most probable path in the diagram.
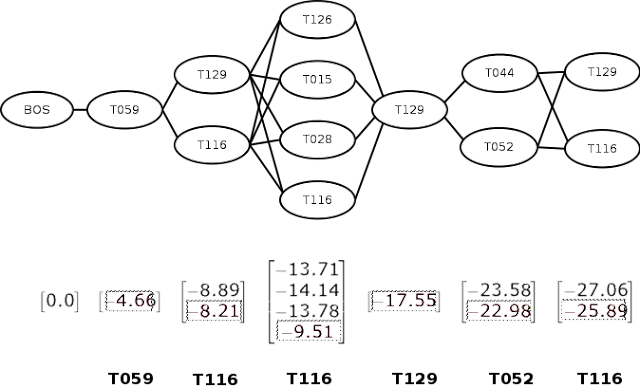
Viterbi algorithm consists of two phases – forward and backward. In the previous phase, we move from left to right, calculating the log-probability of each transition at each step, as shown in the figure by the vectors below each position. When calculating a transition from multiple nodes to a single node (such as from node [T129, T116] that [T126]We calculate both ways and choose the maximum value.
In the backtracking phase, we move from right to left, selecting the maximum likelihood node at each step. These are shown in the figure as boxed entries. We can then search for the corresponding semantic type and return the most likely sequence of semantic types (shown in bold at the bottom of the figure).
However, our goal is to return ambiguous concept associations for entities. Given the uncertain semantic type and multiple possibilities indicated by SciSpacy’s linking process, we use emission probabilities to select the most likely concept that applies to the position. The result of our example is shown in the table below.
| Subject-ID | unit span | Concept-ID | The basic name of the concept | Semantic type code | Semantic type name | it is right? |
|---|---|---|---|---|---|---|
| 1 | coloring | C0487602 | coloring method | T059 | laboratory procedure | N/A* |
| 2 | Antibodies | C0003241 | Antibodies | T116 | amino acid, peptide or protein | Yes |
| 3 | cat | C0008169 | Chloramphenicol O-acetyltransferase | T116 | amino acid, peptide or protein | No |
| 4 | antigens | C0003320 | antigens | T129 | immunological factor | N/A* |
| 5 | Mandatory | C1145667 | Mandatory action | T052 | activity | Yes |
| 6 | Antibodies | C0003241 | Antibodies | T116 | amino acid, peptide or protein | Yes |
(No: unambiguous mappings)
- Code: This github gist contains code that performs NER + NEL on an input sentence using SciSpacy and its UMLS integration, and then uses my adaptation of the Viterbi method (as described in this post ) to resolve ambiguous entity connections.
- Data: I have also provided the transition and emission matrices and their associated lookup tables for convenience, as these can be time consuming to create from scratch from the CORD-19 data set.
As always, I appreciate your feedback. Please let me know if you find any flaws in my approach and/or know of a better approach for unit misunderstandings
[ad_2]
Source link

